The rtemis ecosystem

rtemis provides a comprehensive console and web-based machine learning and visuzalization platform.
- rtemis: R package that provides:
- static and interactive visualization
- unsupervised learning: clustering & decomposition
- supervised learning
- cross-decomposition
- rtemispy: core rtemis API in Python
- Rtemis.jl: core rtemis API in Julia
- rtemislive: web application providing no-code access to rtemis
The Python and Julia ports are currently private and will be made available after the rtemis 1.0 release, aiming for a similar API across languages.
rtemis
The core rtemis API, written in R. rtemis & rtemislive are complementary and interoperable.
For more information on the rtemis package see the online documentation and GitHub repository.

rtemispy
rtemispy is the core rtemis API in Python.

Rtemis.jl
Rtemis.jl is the core rtemis API in Julia.

rtemislive
rtemislive is a comprehensive, modular, and customizable web-based data science platform. It is the graphical user interface for rtemis. It is built using shiny in R.
rtemislive is currently available on the internal UCSF network.
This section provides on an overview of the functionality provided by the package ahead of its public release.

Modules
rtemislive provides access to multiple modules which can be toggled before or after launch of the app.
Data Viewer
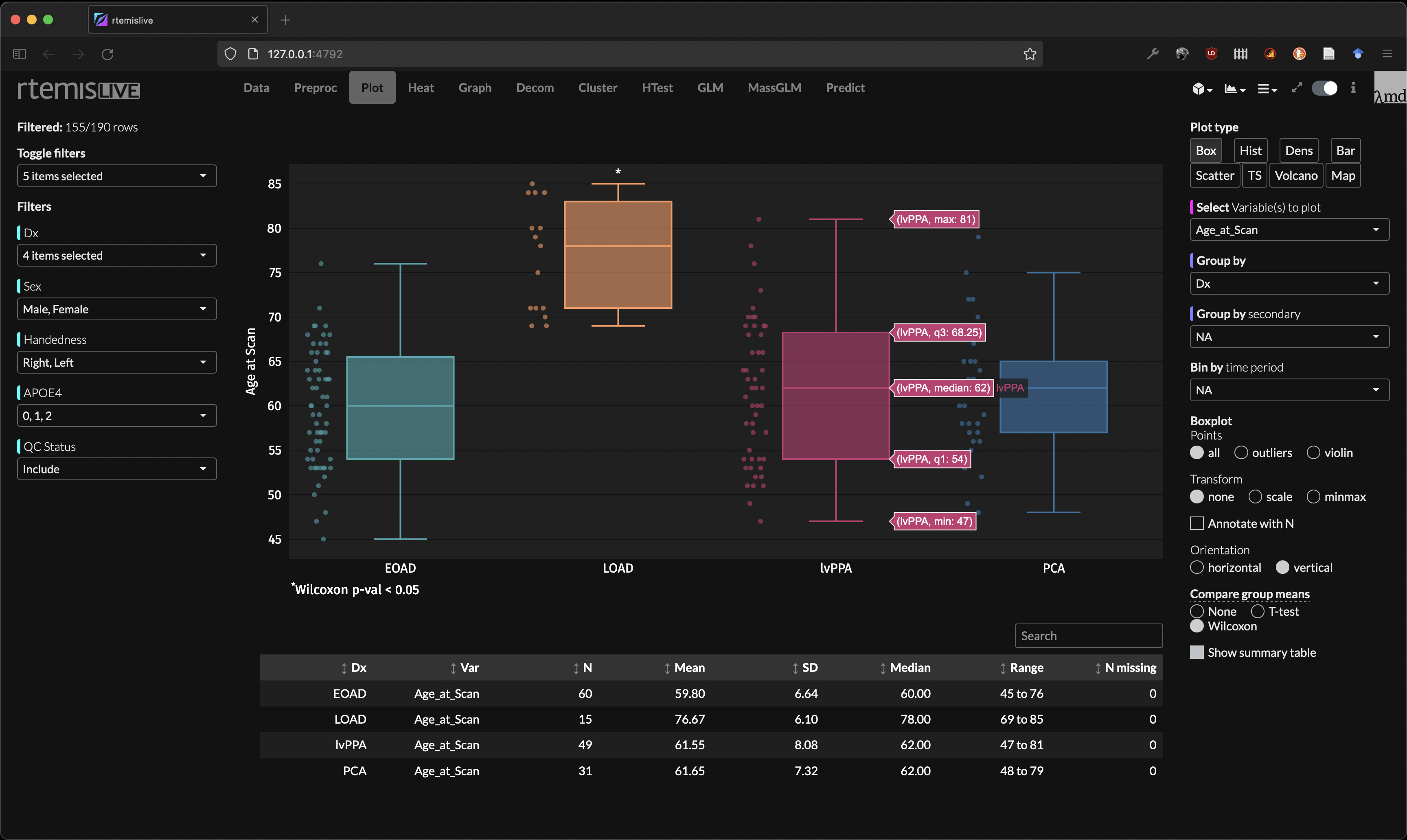
Boxplot
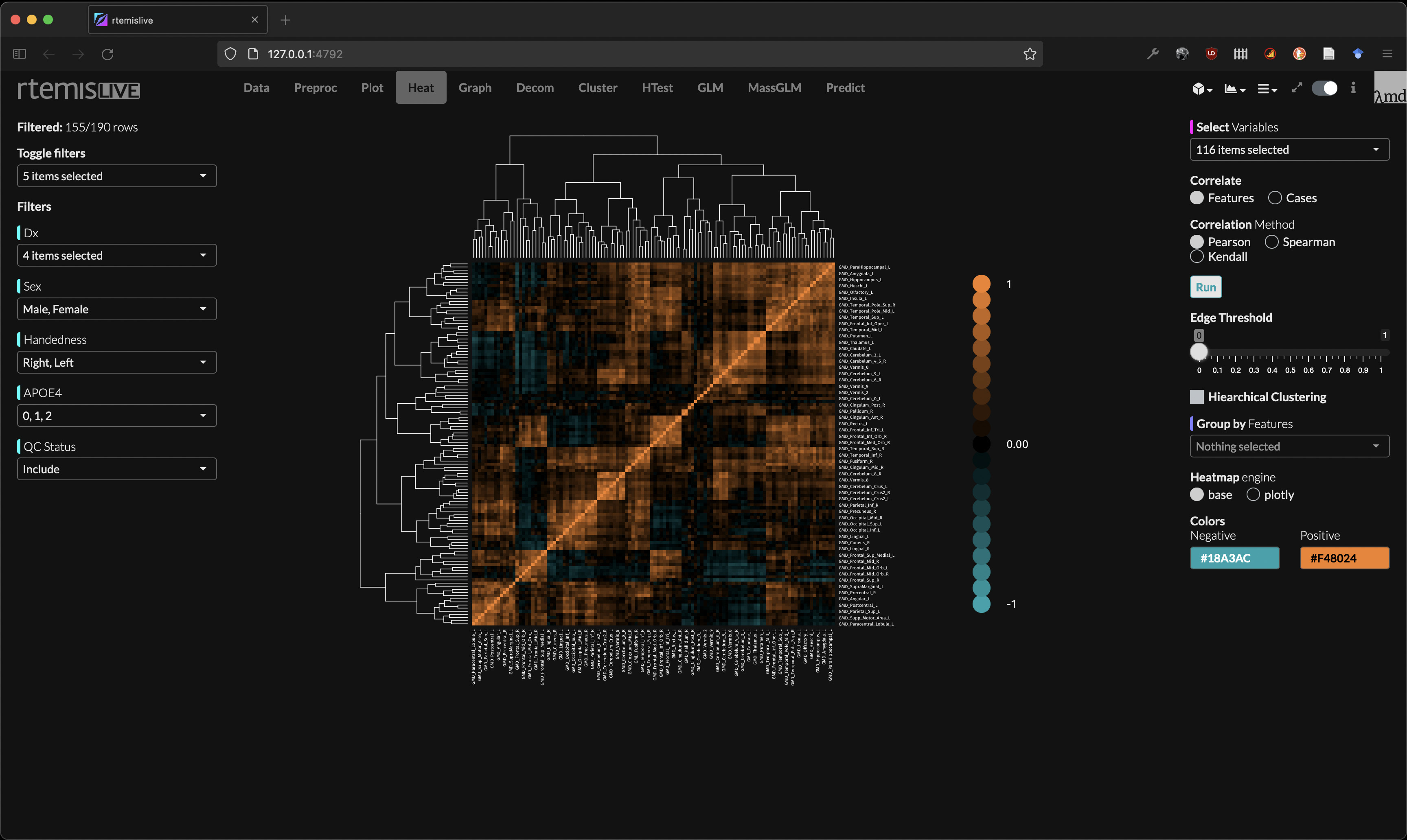
Heatmap
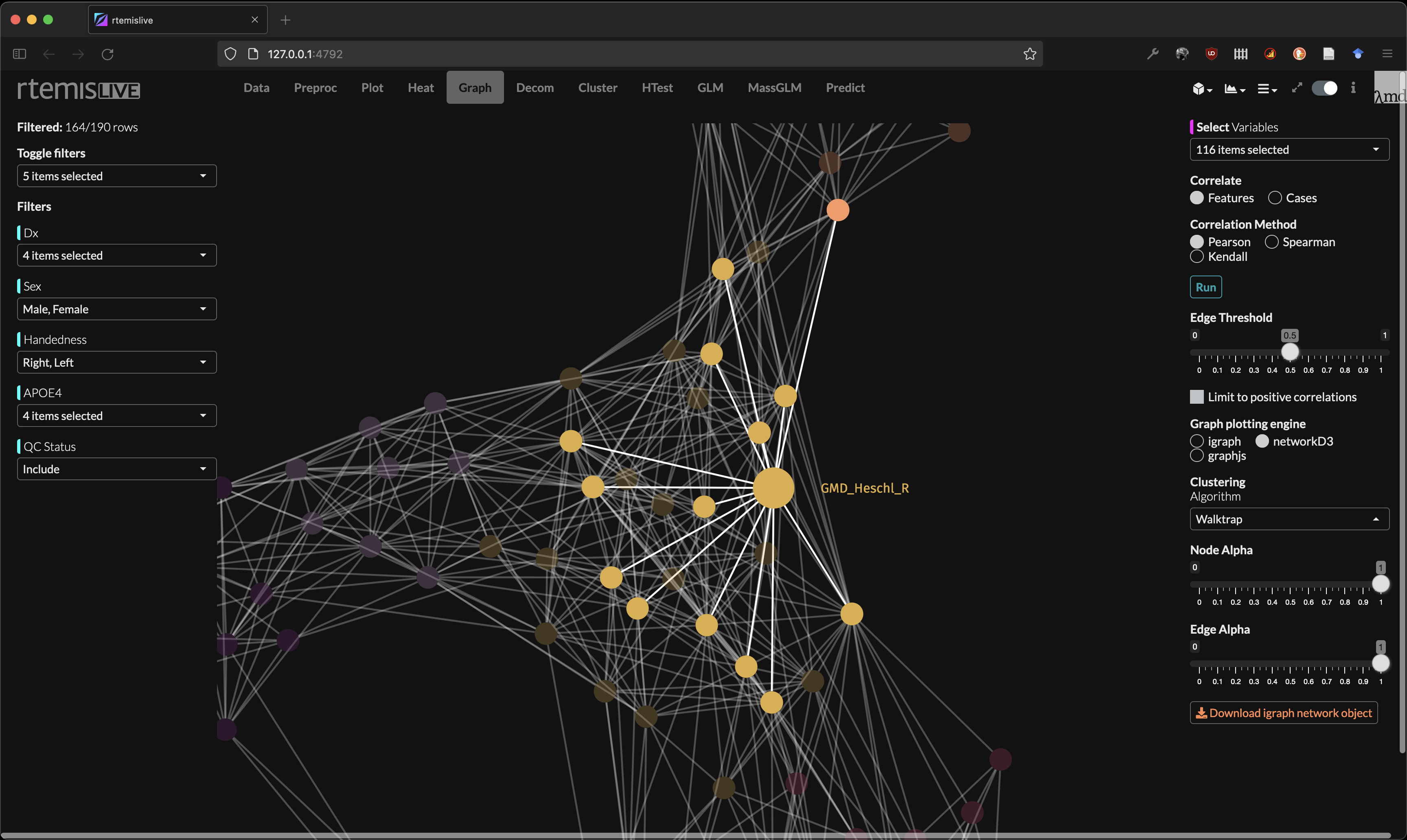
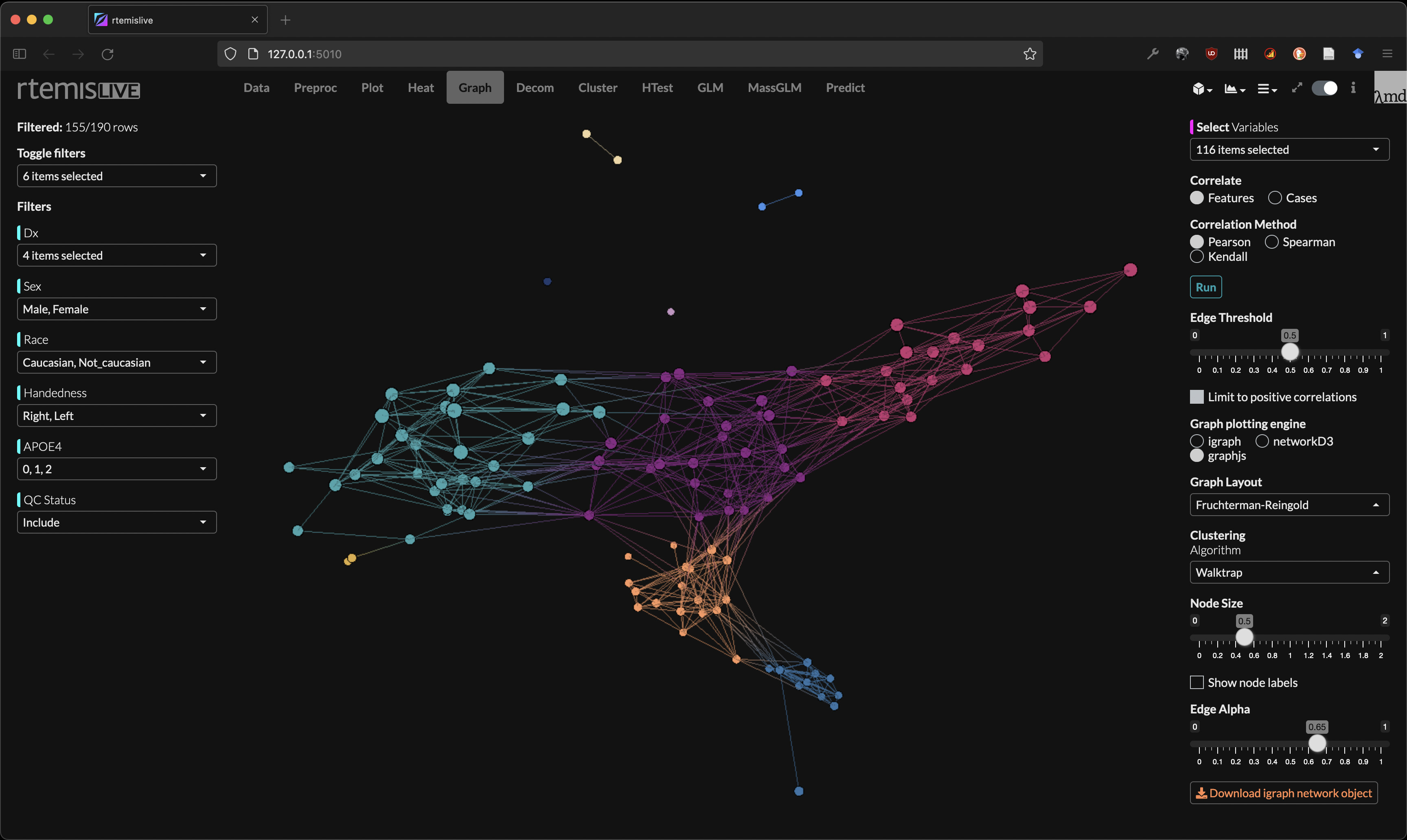
Network analysis
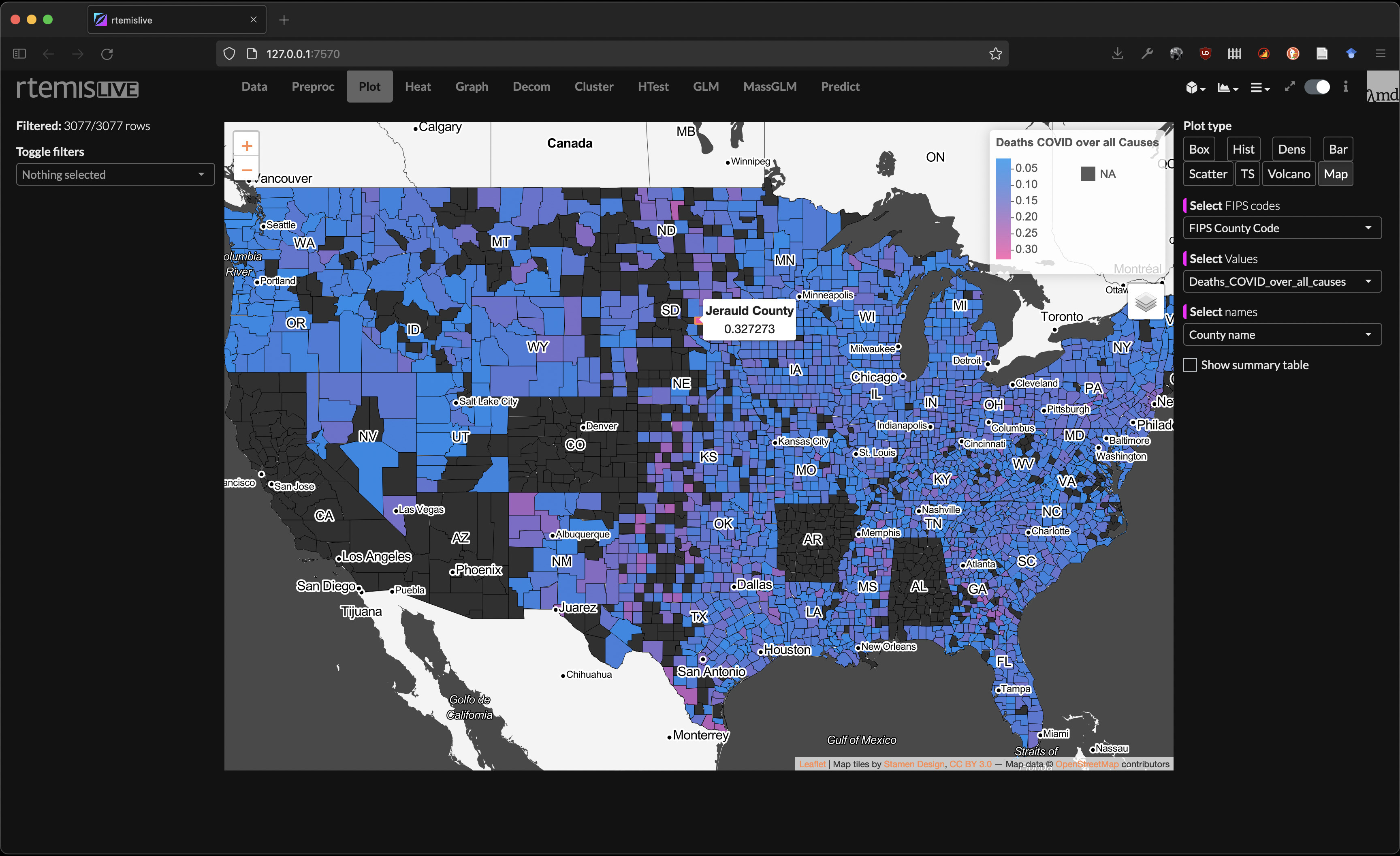
Map Viewer
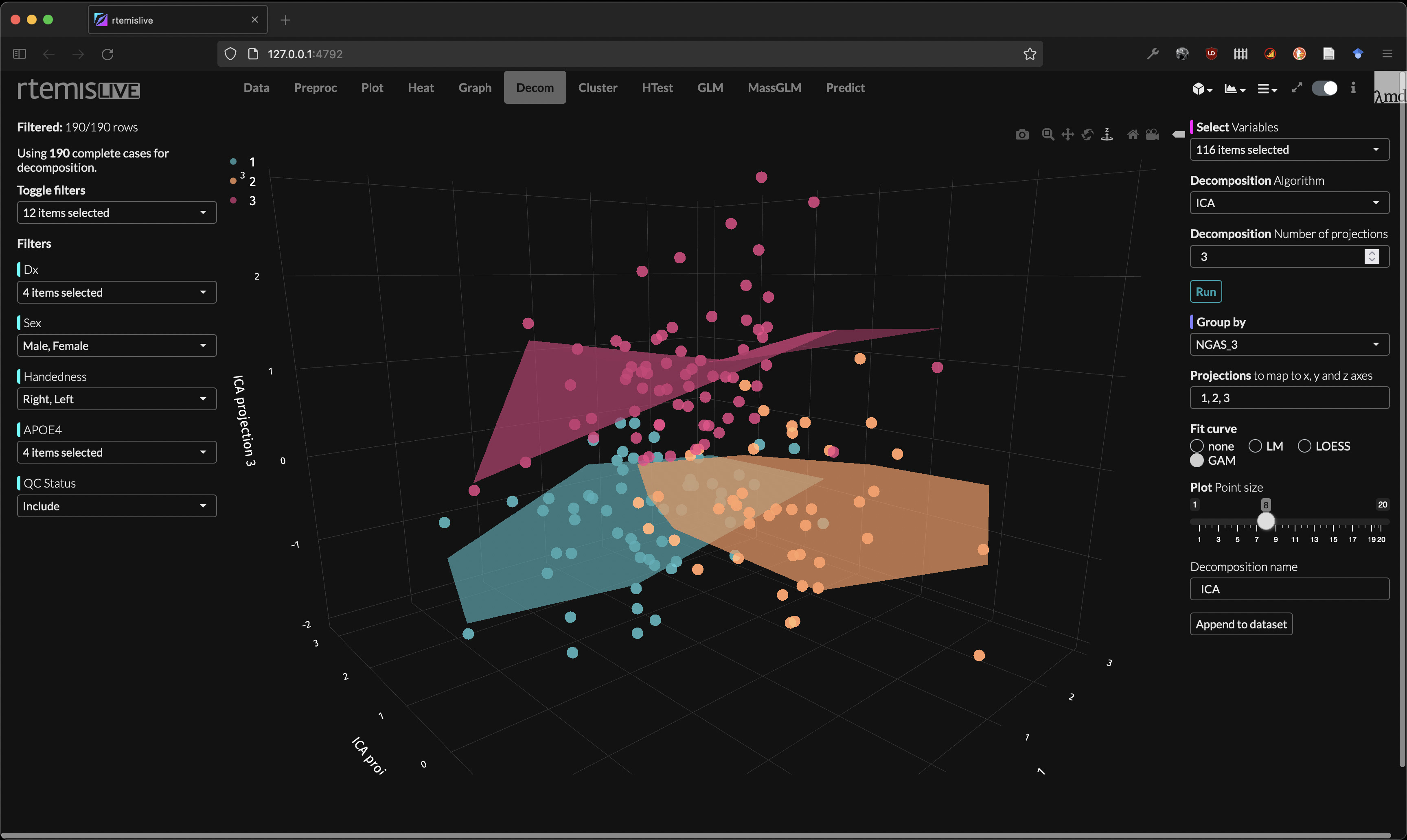
Decomposition
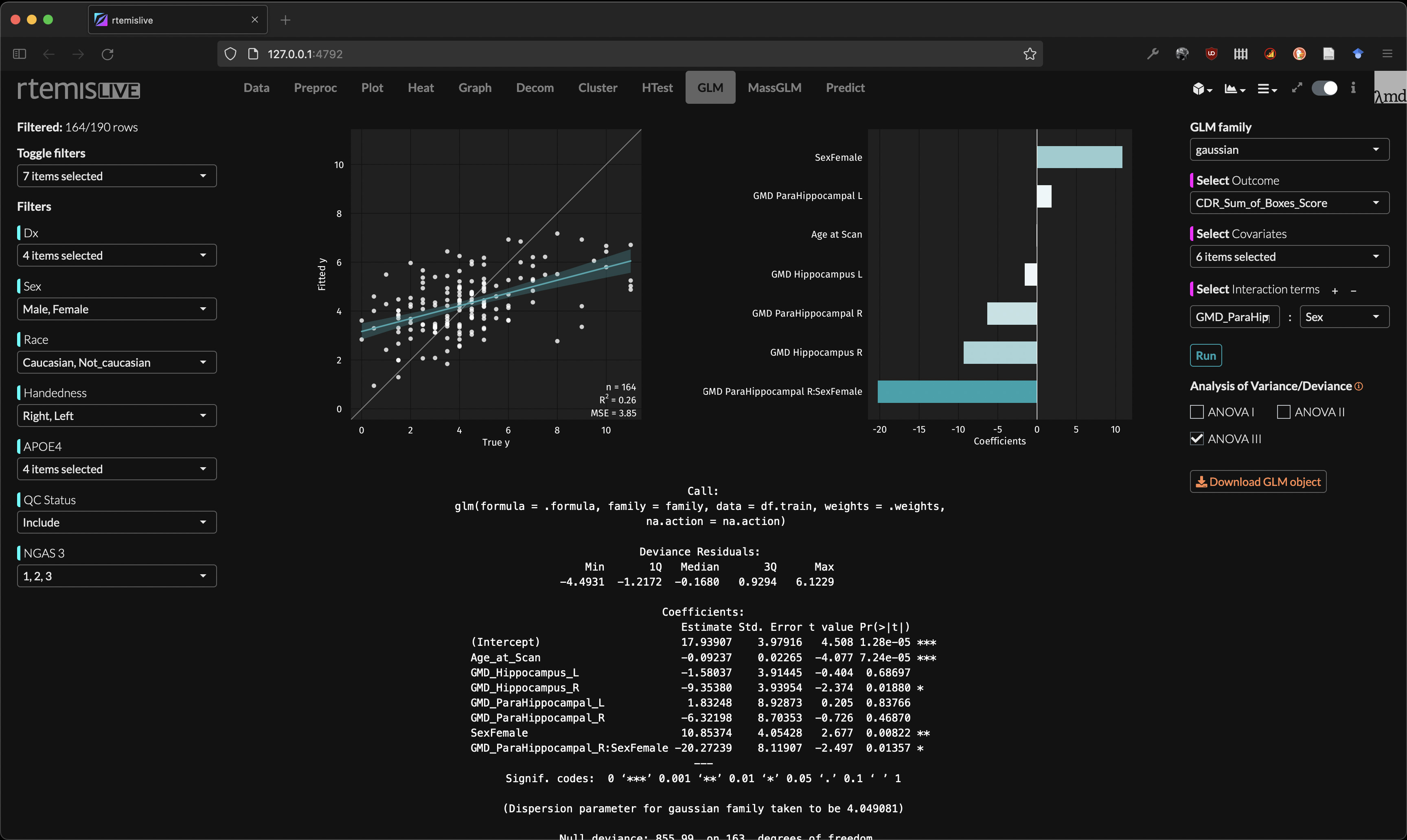
GLM
mass-GLM
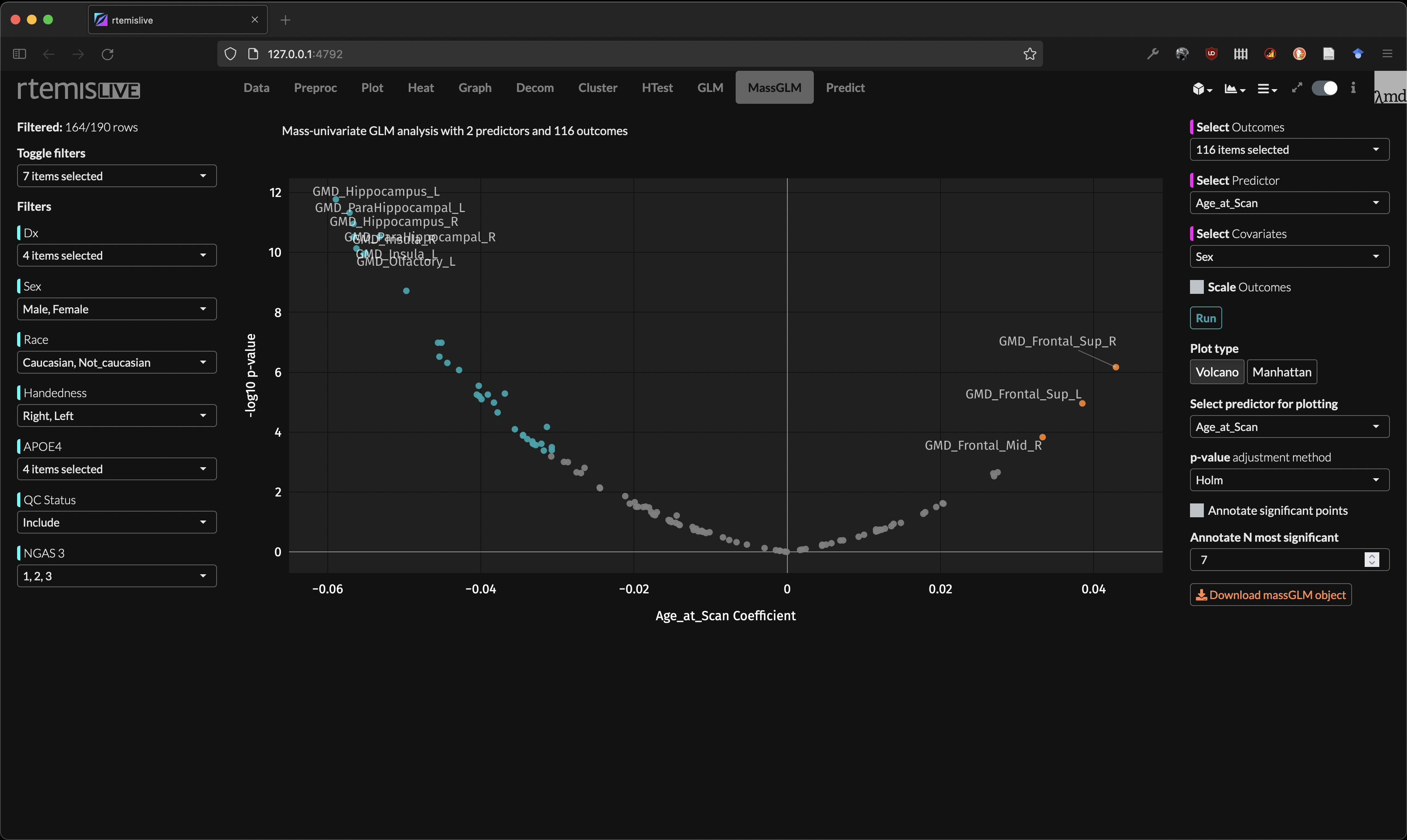
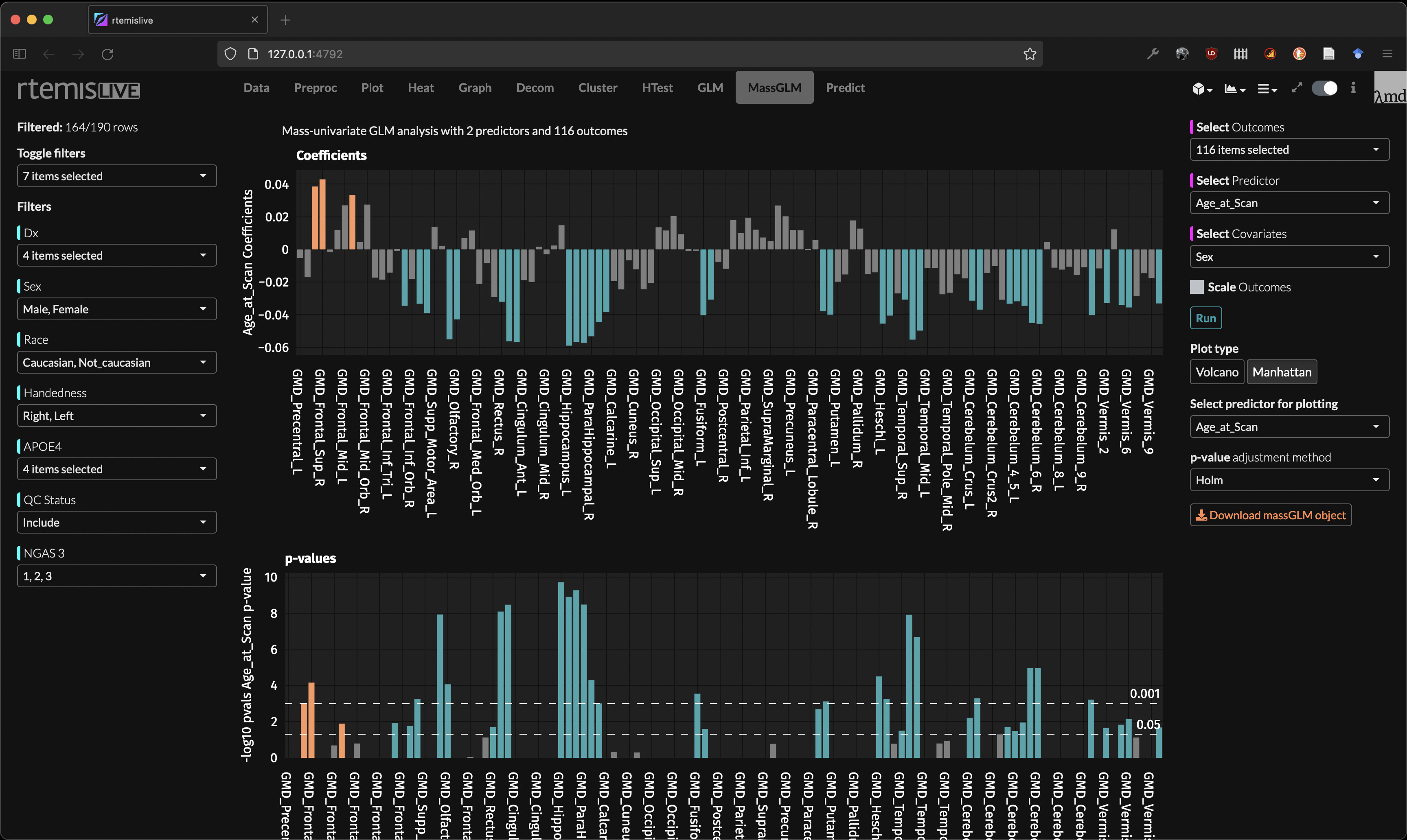
Predictive modeling

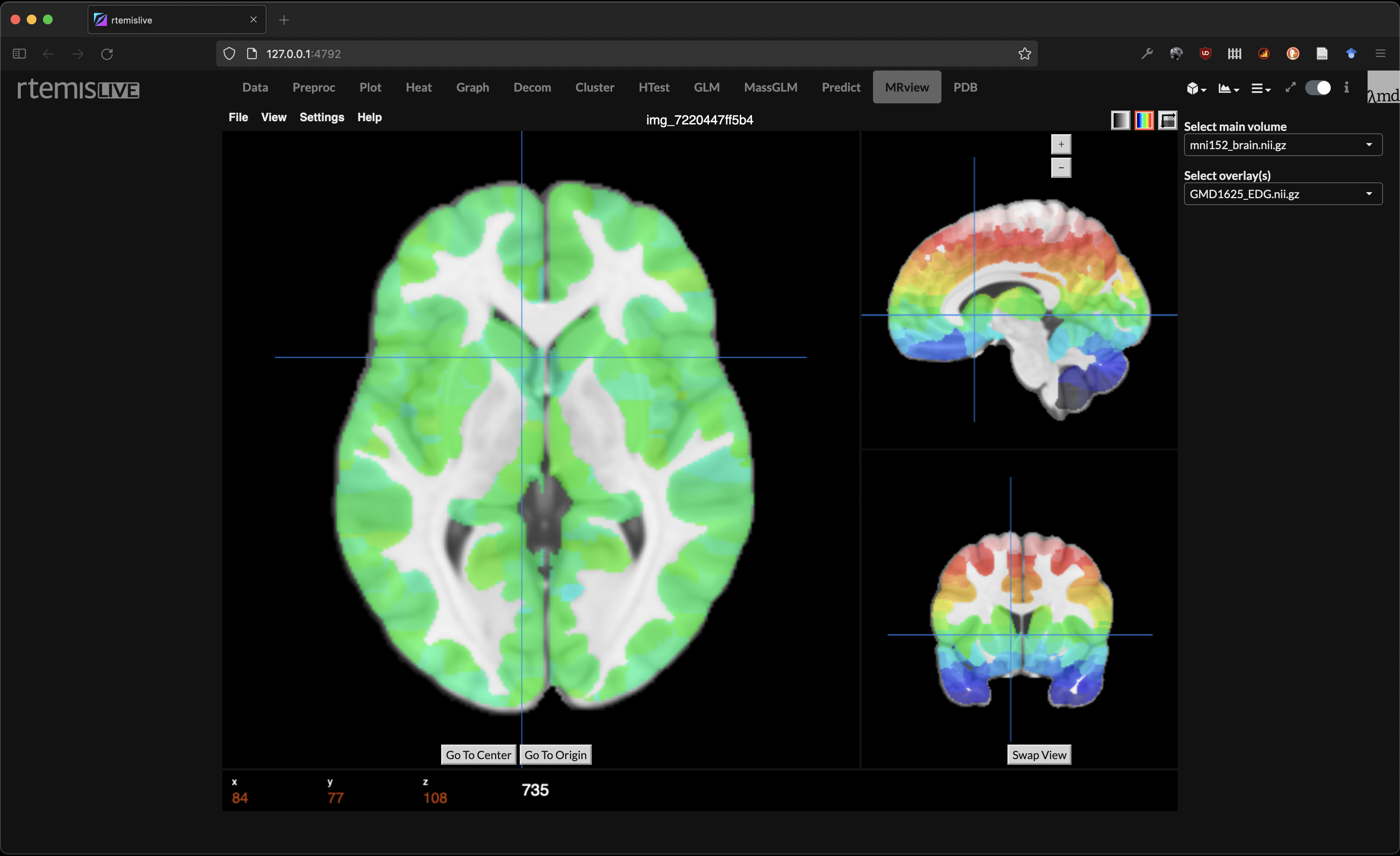
MRI Viewer
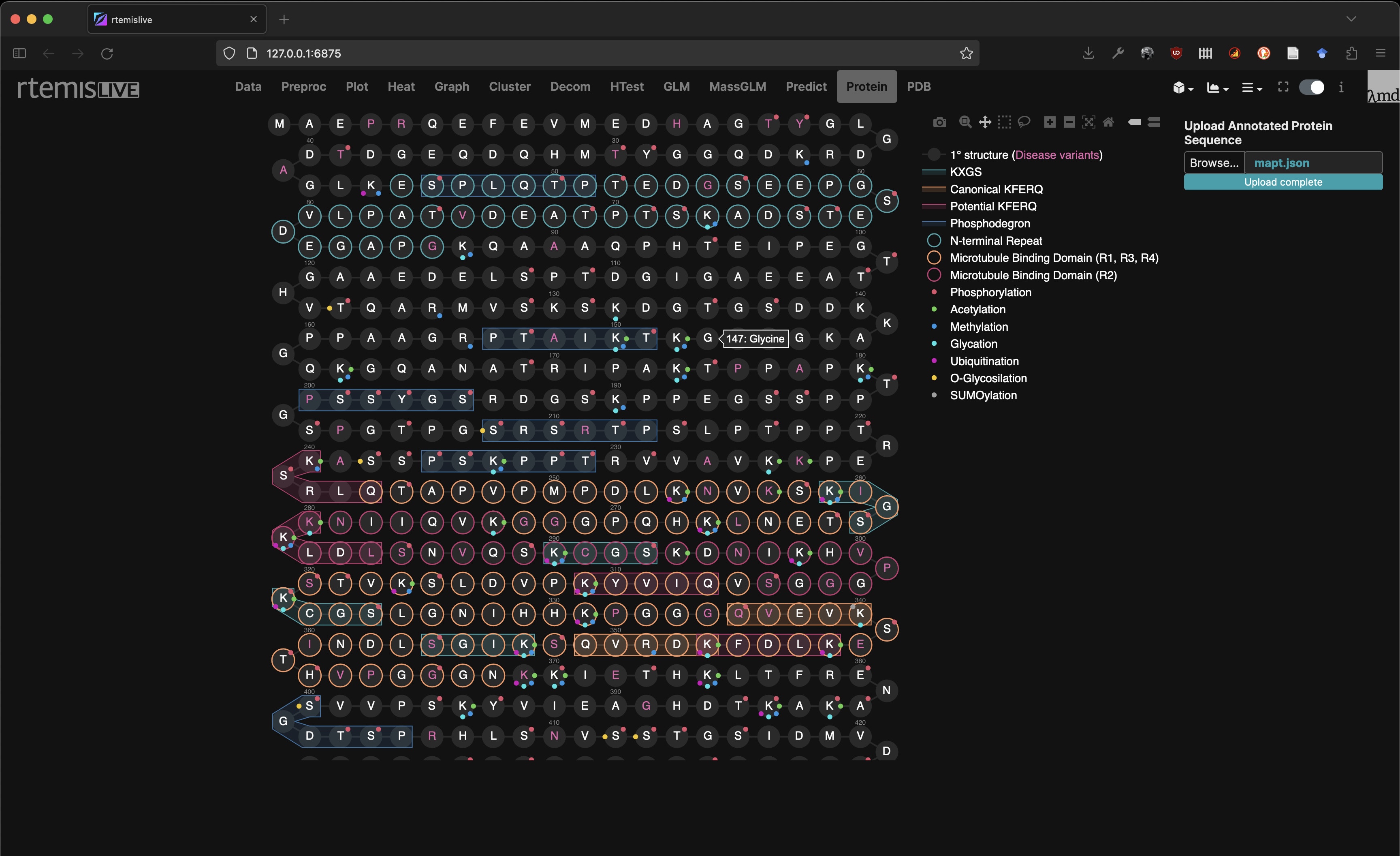
Protein Viewer
PDB Viewer

Built-in help
rtemislive uses tooltips to provide context specific help throughout the UI and also includes builtin help:
List of available modules
- Data
- Preprocessing
- Plot
- Boxplot
- Histogram
- Density
- Bar plot
- Scatter
- Timeseries
- Volcano
- Map
- Heatmap
- Graph (network) visualization
- Survival
- Data Key
- Table 1
- Decomposition / Dimensionality reduction
- Clustering
- Hypothesis Testing
- Generalized Linear Models (GLM)
- mass-GLM
- Predictive Modeling
- GLMNET
- SVM
- CART
- Random Forest
- Gradient Boosting
- XGBoost
rtemisbio
rtemisbio is a new package, that forms the bionformatics extension of rtemis. It is being developed to suppor the mission of the Genomics and Transcriptomics Core of the FTD Center Without Walls.

rtemis SeqVizLive
SeqVizLive, is a web app being developed to provide interactive visualization of annotated genomics and proteomics sequence data. It is as a shinylive app, i.e. a shiny app that uses WebR to run entirely in the user’s browser, without the need for a dedicated shiny server. It is built on top of rtemis and rtemisbio. The initial release is available here.
